SeqNLS: Nuclear Localization Signal Prediction Based on Frequent Pattern Mining and Linear Motif Scoring | PLOS ONE

Nuclear Localization Signal | In Silico Nuclear Localization Signal Prediction | NLS Mapper| - YouTube

NLStradamus: a simple Hidden Markov Model for nuclear localization signal prediction | BMC Bioinformatics | Full Text

DNA binding residues in region F1 acts as a Nuclear Localization Signal... | Download Scientific Diagram

Figure 2.1 from Nuclear export signals (NESs) in Arabidopsis thaliana : development and experimental validation of a prediction tool | Semantic Scholar

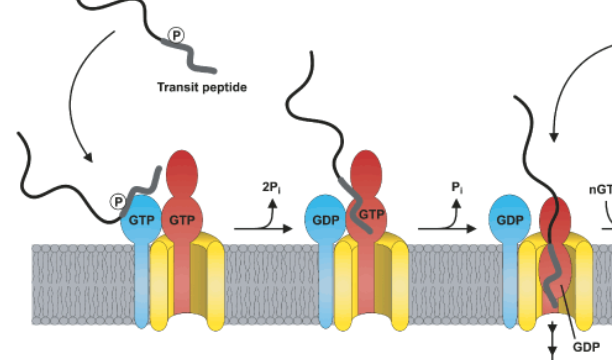
Mapping of NLS and NBLS in Arabidopsis phyB (A) Schematic illustration... | Download Scientific Diagram

Cells | Free Full-Text | Bioinformatics and Functional Analysis of a New Nuclear Localization Sequence of the Influenza A Virus Nucleoprotein

Analysis of rice nuclear-localized seed-expressed proteins and their database (RSNP-DB) | Scientific Reports

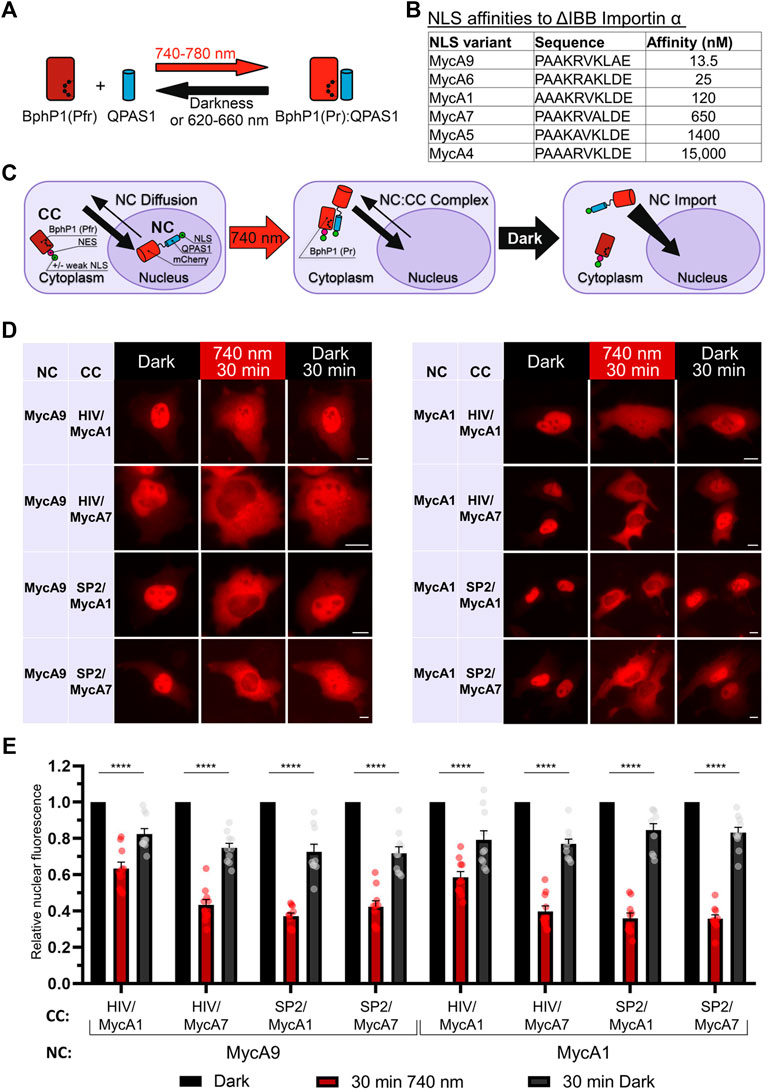
NLS Mapper, PredictProtein and COMPARTMENT OAS3 nuclear localization... | Download Scientific Diagram

Figure 2.3 from Nuclear export signals (NESs) in Arabidopsis thaliana : development and experimental validation of a prediction tool | Semantic Scholar

Identification of a nuclear localization signal in the Plasmodium falciparum CTP: phosphocholine cytidylyltransferase enzyme | Scientific Reports

Analysis of rice nuclear-localized seed-expressed proteins and their database (RSNP-DB) | Scientific Reports

Nuclear Localization Signal | In Silico Nuclear Localization Signal Prediction | NLS Mapper| - YouTube
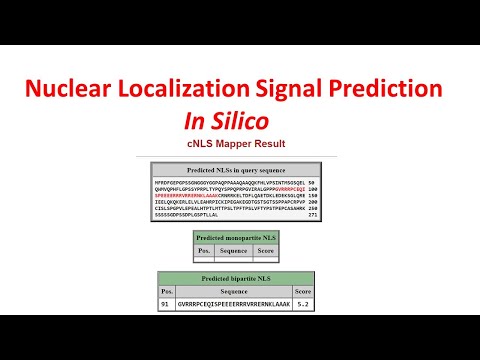
Nuclear Localization Signal | In Silico Nuclear Localization Signal Prediction | NLS Mapper| - YouTube
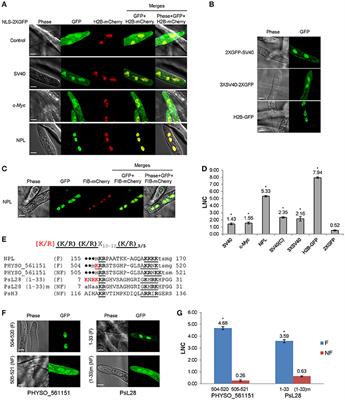
NES and NLS prediction in the bHLH domain of TCF4. (A) Results of NES... | Download Scientific Diagram

Prediction results for putative NLS, NES and DNA-binding motifs on CAV... | Download Scientific Diagram
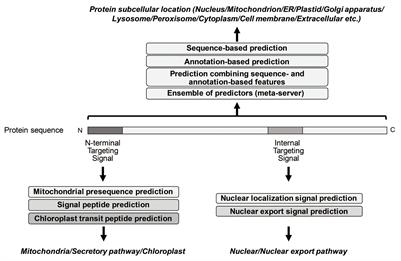
Frontiers | Tools for the Recognition of Sorting Signals and the Prediction of Subcellular Localization of Proteins From Their Amino Acid Sequences

Software tools for simultaneous data visualization and T cell epitopes and disorder prediction in proteins - ScienceDirect
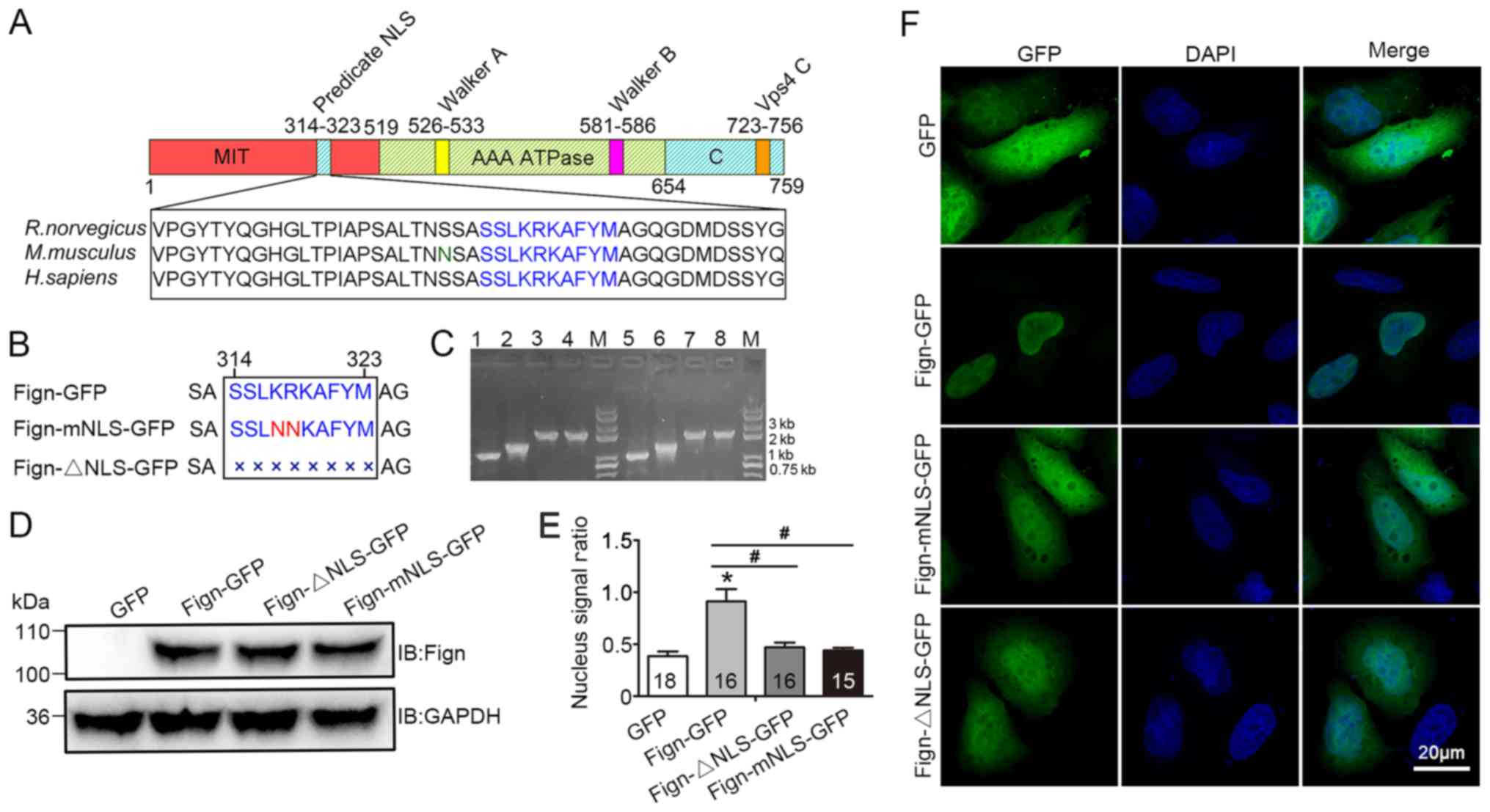
A nuclear localization signal is required for the nuclear translocation of Fign and its microtubule‑severing function
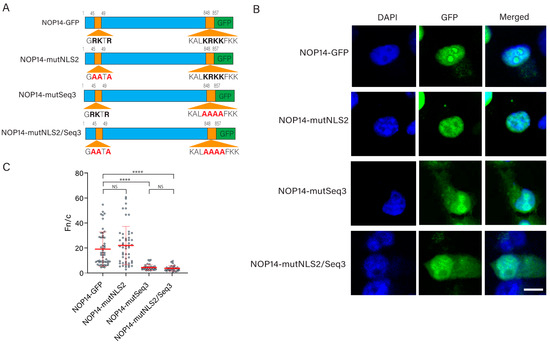
Cells | Free Full-Text | Bioinformatics and Functional Analysis of a New Nuclear Localization Sequence of the Influenza A Virus Nucleoprotein

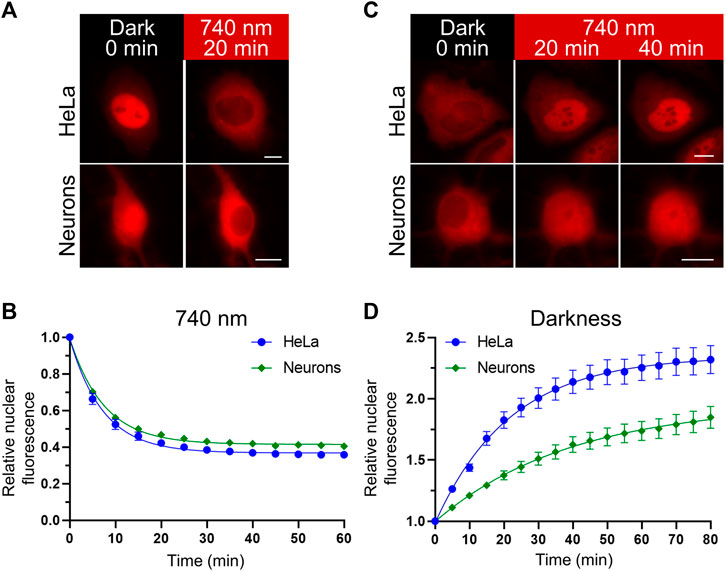
A Novel Proteomic Method Reveals NLS Tagging of T-DM1 Contravenes Classical Nuclear Transport in a Model of HER2-Positive Breast Cancer: Molecular Therapy Methods & Clinical Development

Delivery of eGFP fused with nuclear localization signals (NLS) to cells... | Download Scientific Diagram